The identification and characterization of KNOX gene family as an active regulator of leaf development in Trifolium repens
2022年10月04日 19:34
Jinwan Fan †, Gang Nie †, Jieyu Ma, Ruchang Hu, Jie He, Feifei Wu, Zhongfu Yang, Sainan Ma, Xin Zhang and Xinquan Zhang *
* Correspondence: zhangxq@sicau.edu.cn
† These authors contributed equally to this work.
DOI: 10.3390/genes13101778
发表期刊:Genes
链接:https://www.mdpi.com/2073-4425/13/10/1778
Abstract:Leaf is the primary and critical feed for herbivores. It directly determines the yield and quality of legume forage.Trifolium repens(T. repens) is an indispensable legume species, widely cultivated in temperate pastures due to its nutritional value and nitrogen fixation. Although the leaves ofT. repensare typical trifoliate, they have unusual patterns to adapt to herbivore feeding. The number of leaflets inT. repensaffects its production and utilization. TheKNOXgene family encodes transcriptional regulators that are vital in regulating and developing leaves. Identification and characterization ofTrKNOXgene family as an active regulator of leaf development inT. repenswere studied. A total of 21TrKNOXgenes were identified from the T. repens genome database and classified into three subgroups (Class I, Class II, and Class M) based on phylogenetic analysis. Nineteen of the genes identified had four conserved domains, except forKNOX5andKNOX9which belong to Class M. Varying expression levels of TrKNOX genes were observed at different developmental stages and complexity of leaves.KNOX9, was observed to upregulate the leaf complexity of T. repens. Research onTrKNOXgenes could be novel and further assist in exploring their functions and cultivating high-qualityT. repensvarieties.
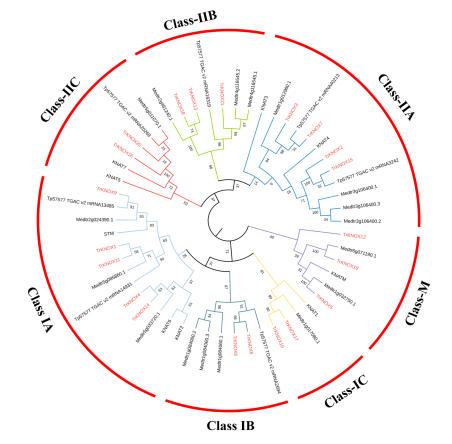
Figure 4.Phylogenetic relationships and motif compositions of KNOX family proteins of four related species. The tree divided the TrKNOX proteins into three subgroups, Class I, Class II, and Class M.